Overview:
In this short article, I am going to show how sentence transformers can be used to create embeddings of text and how to use DBSCAN to detect clusters.
Install Dependencies:
Install the below dependencies,
!pip install -U sentence-transformers
!pip install sklearn
!pip install plotly
!pip install nbformat>=4.2.0
!pip install matplotlib
Import Dependencies:
import pandas as pd
import numpy as np
import plotly.graph_objects as go
import pickle
import matplotlib.pyplot as plt
from sentence_transformers import SentenceTransformer, util
from sklearn.neighbors import NearestNeighbors
from sklearn.cluster import DBSCAN
Data Cleaning:
I was using some custom data which had many inconsistencies. You can create your script for data cleaning. Mine is as below,
def data_cleaning(input_file, output_file):
list_data = ["<br/>","Hi team -","hi team -","Hello-","Hi Team,","Hi team,","Hello,","Hi Team:","Hello Team,","Hi,","Hi team-",
"Hi Team-","Hi Team.","Hello,","hello,","Hi Team","hi team","hello team","Hello!","team,","Team","====","HI","Hi team"]
with open(input_file, 'r') as file:
data = file.read()
for l_data in list_data:
data = data.replace(l_data, "")
# Opening our text file in write only mode to write the replaced content
with open(output_file, 'w') as file:
file.write(data)
df = pd.read_csv(output_file)
# Renaming the column
df.rename(columns={"First Comment": "comments"}, inplace=True)
# Removing blank rows
df.dropna(inplace=True)
df.reset_index(drop=True, inplace=True)
df.to_csv(output_file, index=False)
number_of_rows_input_file = pd.read_csv(input_file).shape[0]
number_of_rows_output_file = pd.read_csv(output_file).shape[0]
print(f"Number of rows in {input_file} = {number_of_rows_input_file}\n" +
f"Number of rows in {output_file} = {number_of_rows_output_file}\n" +
f"Number of rows dropped = {number_of_rows_input_file - number_of_rows_output_file}\n")
print(f"Data cleaning complete. Cleaned data is stored in {output_file}")
# Call the function
data_cleaning("./Data/Raw_Dataset.csv", "./Data/Cleaned_Dataset.csv")
Convert the cleaned dataset to a list:
We need to convert the cleaned dataset to a list to use in sentence transformers for creating embeddings.
# Converting the cleaned dataset into list
df1 = pd.read_csv("./Data/Cleaned_Dataset.csv")
comments_list = df1.comments.tolist()
Model to create embeddings:
We are using sentence transformers to create embeddings.
You can use any model that is supported by sentence transformers. I am using all-MiniLM-L6-v2.
distilbert-base-nli-stsb-quora-ranking is also another good alternative.
def create_and_store_embeddings() -> np.ndarray:
model = SentenceTransformer('all-MiniLM-L6-v2')
embeddings = model.encode(comments_list, show_progress_bar=True)
# Normalize the embeddings to unit length
embeddings = embeddings / np.linalg.norm(embeddings, axis=1, keepdims=True)
return embeddings
# Call the function
create_and_store_embeddings()
Storing the embeddings:
Store the embeddings so that we don't have to create them every time.
# Store sentences and embeddings to disc
with open("./Data/embeddings.pkl", "wb") as fOut:
pickle.dump({'sentences': comments_list, 'embeddings': embeddings}, fOut, protocol=pickle.HIGHEST_PROTOCOL)
Loading the stored embeddings:
Load the embeddings from the already created pickle file as below,
#Load sentences & embeddings from disc
embeddings = np.empty_like(embeddings)
with open("./Data/embeddings.pkl", "rb") as fIn:
stored_data = pickle.load(fIn)
stored_sentences = stored_data['sentences']
embeddings = stored_data['embeddings']
Detect Clusters:
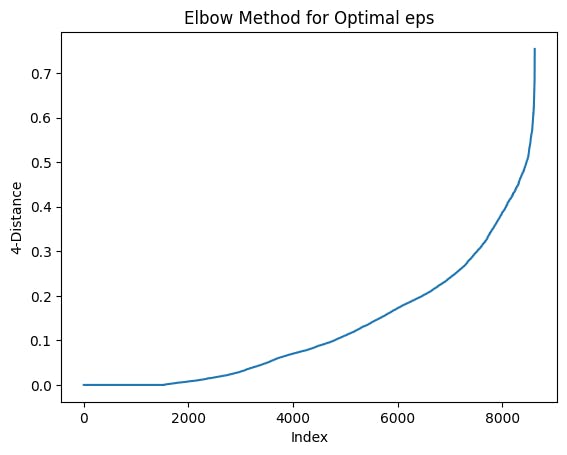
Elbow Method:
To detect the clusters using DBSCAN, we need to find the eps value.
This value can be found by using the Elbow Method. The flow of the method is defined below,
Compute the k-distance graph: Calculate the distance between each point and its kth nearest neighbor (e.g., k=4).
Sort the distances in ascending order and plot them.
Look for the "elbow point" in the plot, which is the point where the curve starts to bend or the rate of change in distances decreases significantly.
Use the distance value at the elbow point as the eps value for DBSCAN.
def find_optimal_eps(embeddings, k=4):
neighbors = NearestNeighbors(n_neighbors=k+1, metric='cosine')
neighbors_fit = neighbors.fit(embeddings)
distances, _ = neighbors_fit.kneighbors(embeddings)
sorted_distances = np.sort(distances[:, -1], axis=0)
plt.plot(sorted_distances)
plt.xlabel('Index')
plt.ylabel(f'{k}-Distance')
plt.title(f'Elbow Method for Optimal eps')
plt.show()
# Call the function
find_optimal_eps(embeddings, k=4)
Example visualization created using Elbow Method:
Cluster Detection using DBSCAN:
DBSCAN is one example of a model to create clusters. K-means is also another good alternative.
The eps parameter controls the maximum distance between two points for them to be considered part of the same neighborhood (i.e., cluster).
The min_samples parameter determines the minimum number of points required to form a dense region, effectively determining the minimum size of a cluster.
You can try different values for eps and min_samples based on your dataset.
def detect_clusters_dbscan(embeddings, eps=0.2, min_samples=10):
dbscan = DBSCAN(eps=eps, min_samples=min_samples, metric='cosine')
cluster_labels = dbscan.fit_predict(embeddings)
unique_communities = []
for label in np.unique(cluster_labels):
cluster_indices = np.where(cluster_labels == label)[0]
if len(cluster_indices) >= min_samples:
unique_communities.append(cluster_indices.tolist())
return unique_communities
# Call the function
uniques_comm = detect_clusters_dbscan(embeddings, eps=0.3, min_samples=30)
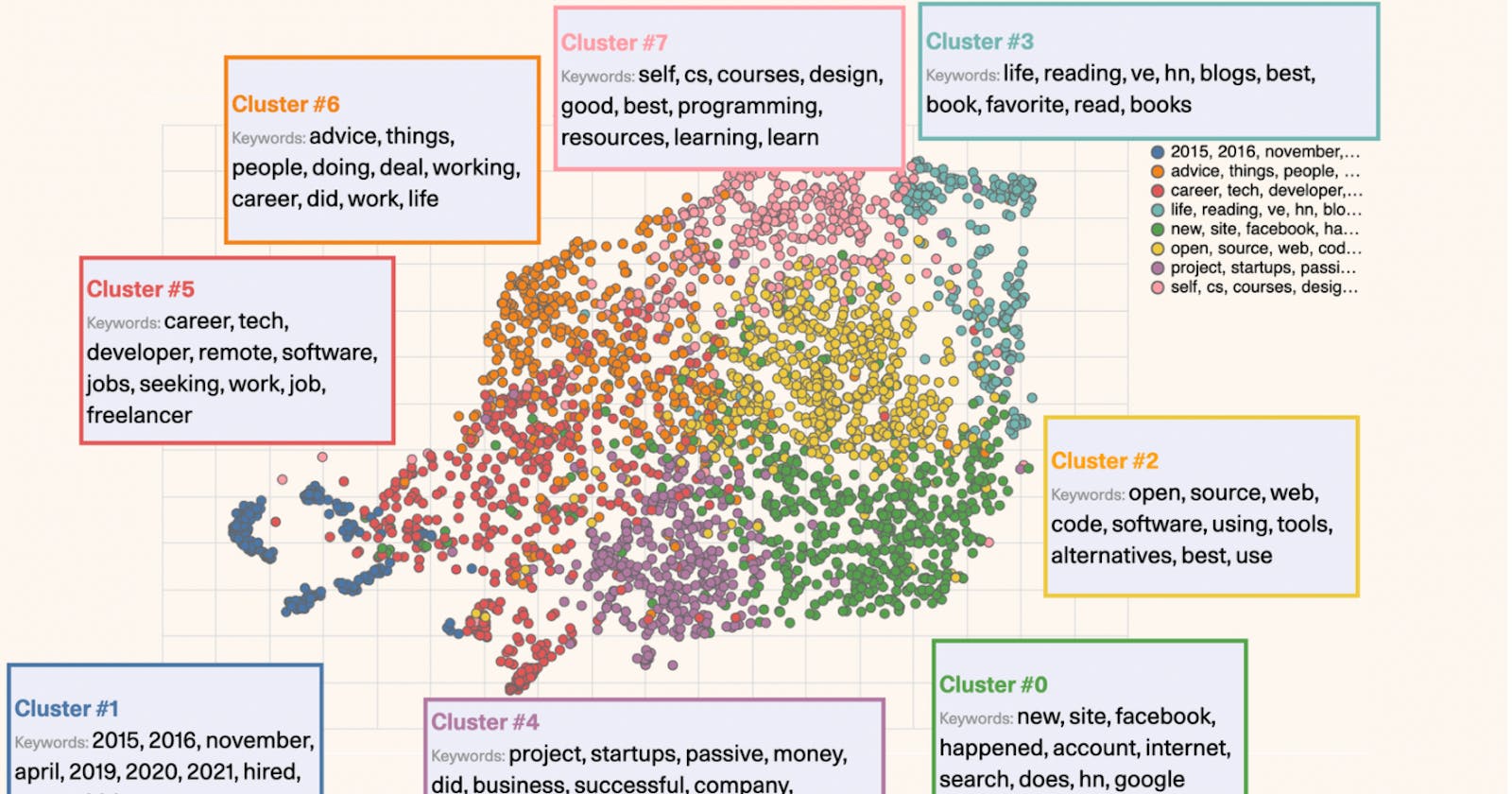
Plot Clusters:
To make it easier to evaluate, we can plot the detected clusters.
def plot_clusters(clusters_to_show):
NUM_CLUSTERS_TO_USE = len(clusters_to_show)
print(f"Number of clusters to use: {NUM_CLUSTERS_TO_USE}")
# if NUM_CLUSTERS_TO_USE > 20:
# NUM_CLUSTERS_TO_USE = 20
sum = 0
for cluster in clusters_to_show[:NUM_CLUSTERS_TO_USE]:
sum += len(cluster)
percentages = []
for cluster in clusters_to_show:
percentages.append((len(cluster)/sum)*100.0)
labels = [f"Cluster{i}" for i in range(1, NUM_CLUSTERS_TO_USE)]
values = percentages
fig = go.Figure(data=[go.Pie(labels=labels, values=values, hole=.3)])
fig.update_layout(width = 720, height = 720)
return fig
# Call the function
plot_clusters(uniques_comm)
Example visualization created using sample dataset: